Expressed Variants from Single Cell RNA Sequencing
We study how genetic variants are expressed at the single-cell level using scRNA-seq. This work helps reveal cell-to-cell differences, track clonal patterns, and understand how genetic diversity shapes health and disease.
- Tools
-
Mutations in Single Cells
Cancer is driven by mutations and genetic variation, but their effects depend on the cellular context. At the Horvath Lab, we study how variants are expressed, regulated, and acted upon in individual cells, using cutting-edge sequencing, computational tools, and AI models.Tools for Single-Cell Variant Analysis
To uncover how mutations shape cell behavior, we develop and share specialized tools:- scExecute – extracts cell-specific reads to discover expressed variants.
- scReadCounts – quantifies variant and reference alleles per cell for allele-specific expression (ASE).
- scSNViz – interactive platform for visualizing expressed SNVs, isoforms, and allelic imbalance.
These tools transform raw data into insights about how mutations drive cell identity,clonal evolution, and disease progression.
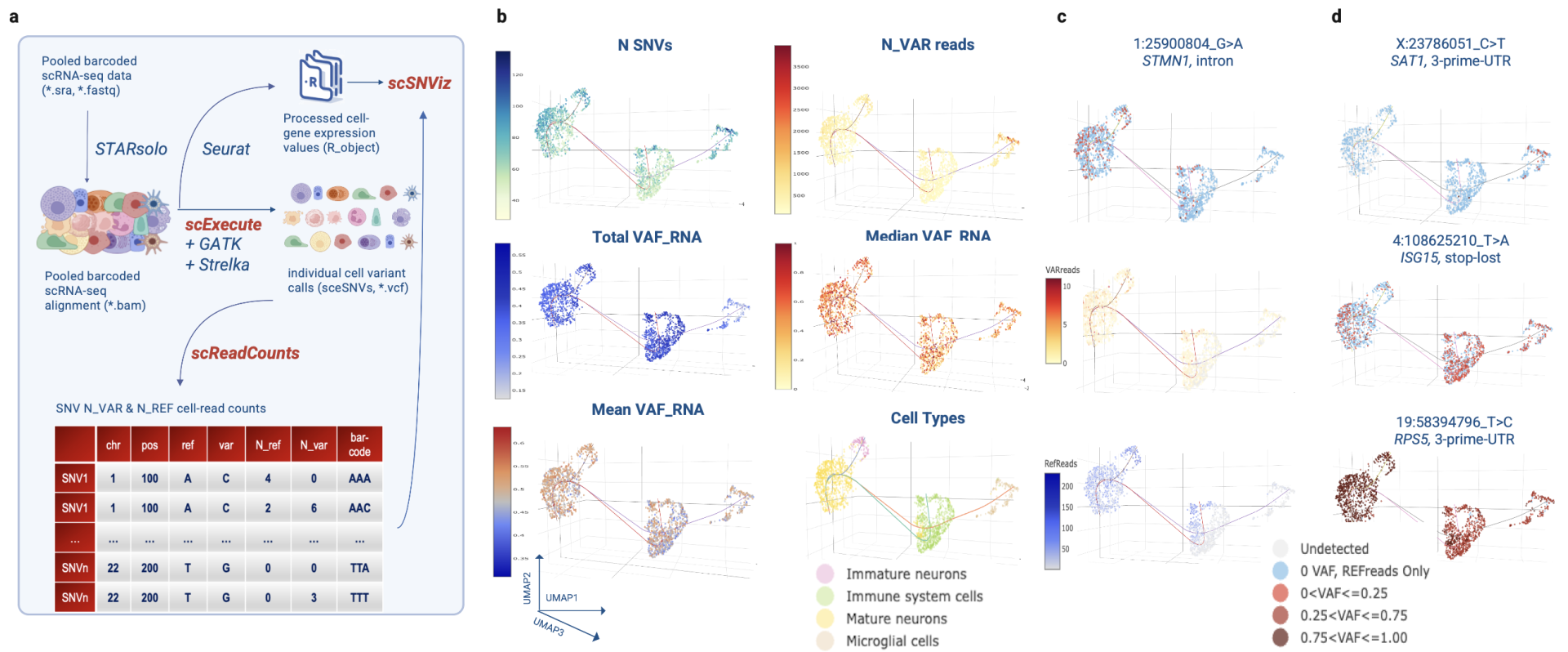
Figure | Single-cell SNV analysis workflow and visualization. Our pipeline extracts cell-specific reads, quantifies variant and reference alleles, and projects them onto transcriptional clusters. This enables discovery of expressed SNVs, mapping of allele-specific expression, and comparison of variant patterns across individual cells and cell types.
- Long-Read Single-Cell Sequencing
-
We apply long-read sequencing (PacBio Revio + 10x Genomics and Oxford Nanopore) to capture the full spectrum of genetic variation in single cells.
- Long reads reveal full-length isoforms, splice variants, fusion transcripts, and RNA modifications, while also detecting within-cell co-dependent features — such as co-occurring allele usage, isoform choice, and expression imbalance — that point to regulatory mechanisms.
- They enable precise mapping of allele-specific expression (ASE) skew, a powerful signal linking genetic variants to their transcriptional impact.
- Importantly, ASE skew provides a functional signature of variant impact. The same mutation may behave differently across cell types (context-specificity), many unannotated variants show strong skew (novel discovery), and ASE skew offers biologically grounded features for AI-driven variant prioritization and therapy response prediction.This work is building the first isoform-aware, cell-resolved maps of genetic variation in tumors and normal tissues — showing that transcriptional imbalance is not noise, but a reproducible readout of how variants shape biology in real time.
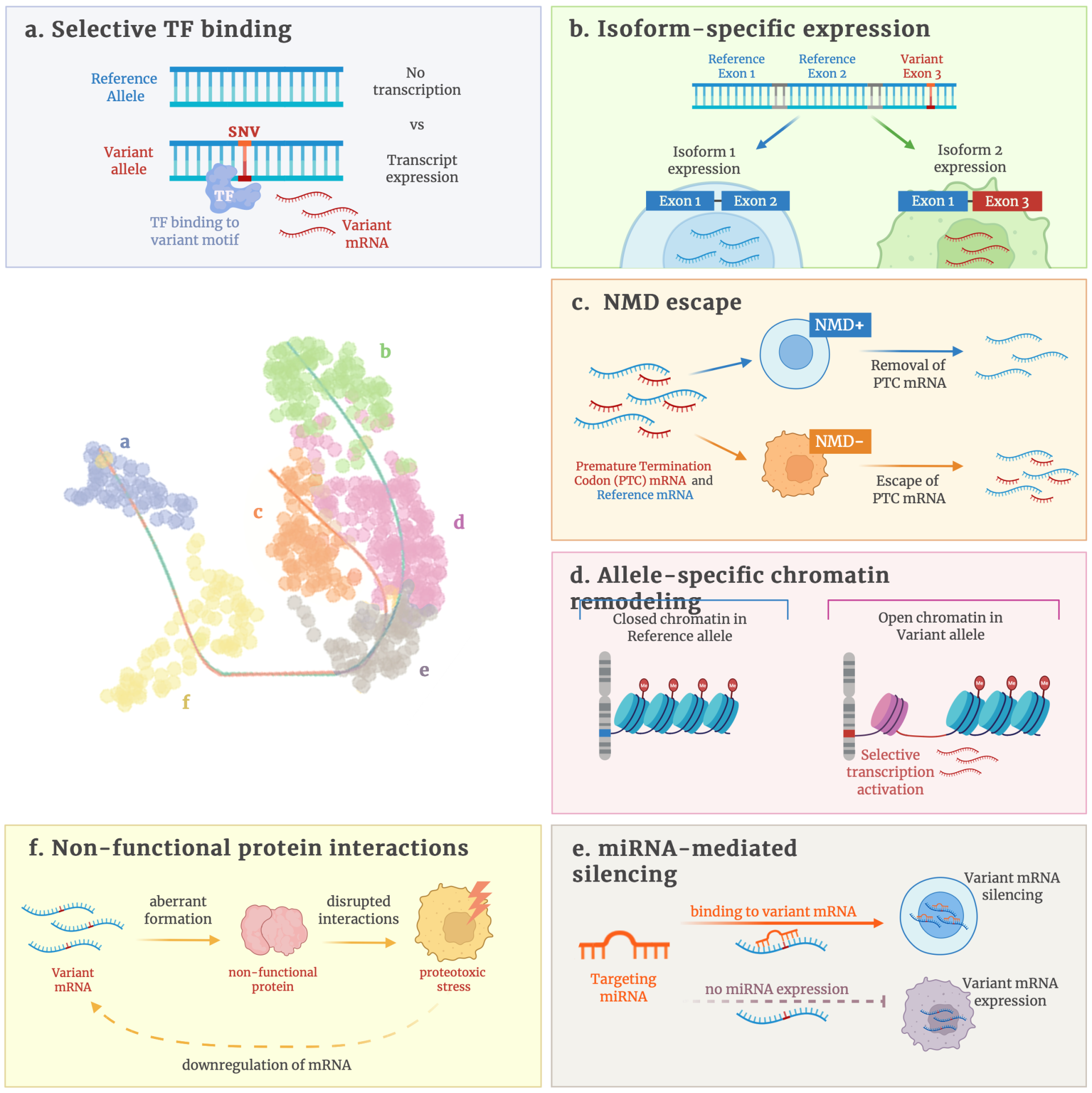
Figure Legend: Mechanisms and single-cell visualization of allele-specific expression skew. (a–f)
Schematic overview of mechanisms through which variants generate cell-type–specific allelic imbalance, including selective TF binding, isoform-specific expression, NMD escape, allele-specific chromatin remodeling, non-functional protein interactions, and miRNA-mediated silencing. (right) Single-cell UMAP projections illustrate variant allele fractions across renal cell populations, highlighting cell-type–restricted expression patterns for specific SNVs (examples: RCC2, ACTG1 3′UTR, SERPINE1 intronic). Together,these panels demonstrate how single-cell long-read sequencing links mutations to regulatory and functional outcomes in defined cellular contexts.
- AI approaches for predicting cell level response to treatment
-
AI Models for Variant Impact
We are developing AI frameworks that integrate single-cell and long-read data to predict variant functionality and therapy response.
- Isoform-aware features: Incorporate full-length transcript information, splice patterns, and fusion events missed by gene-level models.
- Expressed SNVs and ASE signatures: Use allele-specific expression and variant allele fraction dynamics as biologically grounded predictors.
- Clone- and state-level focus: Models aim to capture therapy resistance in persisting subpopulations rather than bulk averages.
- Designed to outperform gene-level predictors: By leveraging richer features, these models provide finer resolution and improved predictive power.
Our goal is to create AI systems that can anticipate resistance before it emerges clinically, offering a pathway toward real-time, adaptive precision oncology.
- Recent Grants
-
Aug 15, 2025
NIH Funded Project on Early Life Stress and Cardiovascular Disease:
Our lab, together with Dr Paul Marvar’s team, is part of a new multi-disciplinary project exploring how early life stress (ELS) contributes to cardiovascular disease (CVD) risk later in life. Clinical studies show that ELS accelerates biological aging and increases susceptibility to neurodegenerative, cardiovascular, and metabolic conditions, with vascular dysfunction emerging as a central factor.Using a preclinical mouse model, this project will investigate how early adversity influences cardiovascular structure, function, and molecular pathways in adulthood. The findings are expected to advance understanding of the long-term health consequences of ELS, uncover potential sex-specific vascular markers, and highlight new therapeutic opportunities for reducing CVD risk
Grant: NIH R21AG091052Sep 13, 2024
Uncovering Epigenetic Drivers of HIV-Associated Neurocognitive Disorders
Our lab is part of an NIH-funded project led by Dr Michael Bukrinsky at George Washington University investigating how epigenetic mechanisms contribute to HIV-
associated neurocognitive disorders (HAND). Despite effective antiretroviral therapy(ART), HAND remains a common comorbidity, driven in part by persistent neuroinflammation whose underlying causes are poorly understood.Recent studies by Dr Bukrinsky’s team demonstrated that extracellular vesicles (EVs) carrying the HIV-1 protein Nef can induce a form of “trained immunity” in monocytes,leaving behind long-lasting pro-inflammatory epigenetic changes. This project will extend those findings to brain-resident myeloid cells and other neural populations,testing whether Nef-induced epigenetic memory sustains inflammation in the central
nervous system even after viral suppression.By combining humanized mouse models, single-cell epigenomic profiling, and patient-derived brain datasets, this research aims to define how viral proteins reshape the brain’s immune landscape and identify lasting epigenetic signatures of HIV infection.The results could provide critical insight into why neuropathology persists in people living with HIV and inform new strategies for mitigating HAND beyond viral suppression.
Grant: NIH R21NS137986Sep 11, 2024
Lab Joins ARPA-H Funded Multi-Institutional Project
Our lab is part of a multi-institutional team, together with Dr Mazumder’s group, that has been awarded funding from the Advanced Research Projects Agency for Health (ARPA-H). The project, titled Federated Ecosystems for Analytics and Standardization Technologies (FEAST), will advance secure digital healthcare and research platforms.By integrating electronic medical records (EMR), multi-omics data, and biomedical knowledge, FEAST will drive the creation of innovative applications at the intersection of precision medicine, biomedical informatics, and translational research.
Award Number: AY2AX000026July 15, 2023
NIH Funds Innovative Study on BRCA1 and Immune Function
Our lab is contributing to an NIH-funded project led by Dr Rong Li at George Washington University that explores a new dimension of BRCA1 biology: its role in immune cells. While BRCA1 research has largely focused on its function in DNA repair within breast epithelial cells, this project investigates how BRCA1 heterozygosity in CD8+ T cells may also shape cancer risk.Preliminary studies show that women with BRCA1 germline mutations have reduced circulating CD8+ T cells, and mouse models confirm that BRCA1 deficiency in these cells weakens antitumor immunity. This suggests that both genomic instability in breast epithelium and impaired immune surveillance contribute to cancer incidence in BRCA1 mutation carriers.By combining expertise in cancer biology, tumor immunology, and transcriptional
regulation, the study will define how BRCA1 enhances T cell-mediated antitumor activity and examine its impact on immunotherapy. This work represents a paradigm shift, expanding BRCA1 research beyond DNA repair to include immune regulation, with the potential to inform novel immune-boosting strategies for breast cancer prevention.
Grant: NIH R01CA282303Aug 1 st , 2022
NIH Awards Project on BACH1 and Cancer Metabolism
Our lab is part of an NIH-funded project led by Dr Jiyoung Lee at George Washington University to investigate how the transcription factor BACH1 regulates lactate catabolism in triple negative breast cancer (TNBC). Tumor metabolic heterogeneity remains a key challenge in therapy, and this work aims to reprogram cancer metabolism to enhance the effectiveness of metabolic inhibitors. Research from Dr Lee’s team has shown that BACH1 suppresses both mitochondrial activity and lactate oxidation, reducing the ability of TNBC cells to metabolize lactate.Targeting BACH1 may shift cancer cells toward greater reliance on mitochondrial respiration, thereby sensitizing tumors to inhibitors of lactate transport and utilization.Through an integrated approach combining transcriptomics, metabolomics, and advanced tumor models, this study will provide critical insights into how lactate fuels cancer cells and how BACH1 can be leveraged as a therapeutic target.
Grant: NIH R37CA270536
April 1, 2024
Exploring Adipocyte PD-L1 in Breast Cancer Immunity
Our lab is part of an NIH-funded project led by Dr Rong Li at George Washington University investigating how adipocytes shape the breast tumor microenvironment
through immune regulation. While PD-L1 and its receptor PD-1 are major targets of immunotherapy, clinical trials in breast cancer have shown only limited benefit,
suggesting that tissue-specific factors may influence therapeutic response. Recent findings from Dr Li’s group identified adipocytes as an unexpected, functionally
significant source of PD-L1 in breast tissue. Adipocyte PD-L1 is upregulated during adipogenesis and obesity-related inflammation, and early studies show that it promotes tumor growth and suppresses immune activity. This project will examine how adipocyte PD-L1 interacts with lipid metabolism pathways, how it regulates cross-talk among adipose, immune, and tumor cells, and how it affects response to immunotherapy in both obese and non-obese settings.By revealing how adipocyte PD-L1 contributes to immune exclusion and tumor progression, this work aims to provide new mechanistic insight into breast cancer biology and inform strategies to improve checkpoint blockade therapies.
Grant: NIH R01CA279566 - Link to GitHub
-
Check the Horvath Lab tools here: Horvath Lab
Why It Matters
Every tumor is a mosaic of diverse cells carrying different mutations. Bulk sequencing averages these signals, masking the rare but critical variants that drive progression and resistance.
By studying mutations in single cells with long-read sequencing, advanced tools, and AI models, we can:
- Detect functionally important variants in their true cellular context.
- Reveal how specific clones adapt and resist therapy.
-
Build predictive models that anticipate resistance before it emerges clinically.
Our research moves the field closer to real-time, personalized cancer treatment.